Navigation auf uzh.ch
Navigation auf uzh.ch
Funding: SNF project 310030_215282 (2023-2027)
People involved: Dr. Paula Bellés Sancho, Dr. Yilei Liu, Martinus Hjorungnes, Romy Leemann
Collaborators: Dr. Christian Ahrens (Agroscope-Molecular Ecology and Swiss Institute of Bioinformatics), Prof. Nicola Zamboni (IMSB, ETHZ), Dr. Euan K James (The James Hutton Institute, Dundee, UK).
The successful interaction between specific rhizobia and host plants lead to the formation of specialized symbiotic organs, called root nodules, in which the bacteria fix atmospheric nitrogen (N2) and make it available to the plant. In exchange, the plant provides carbon and energy sources generated by photosynthesis. A complex series of events, coordinated by both plant and bacterial signals, underlies the development of this symbiotic interaction1. Until recently, rhizobia were thought to belong exclusively to the alpha-subclass of proteobacteria (alpha-rhizobia, isolated first in 1888). This changed in 2001, when two genera belonging to the beta-proteobacteria (Burkholderia and Cupriavidus) were discovered (beta-rhizobia)2. For the legume nodulating strains belonging to the environmental clade of the genus Burkholderia, recently a new genus, Paraburkholderia, has been proposed. The strain Paraburkholderia phymatum STM815T stands out for its ability to establish a nitrogen-fixing symbiosis with more than 50 different legumes (including mimosoid and papilionoid) as well as to reduce nitrogen in free-living conditions (Figure 1a,b)3–8. Moreover, the availability of the complete genome sequence makes it an ideal model to characterize the whole gene/protein expression profile of P. phymatum STM815T grown in different environments including the root nodule5.
In order to understand the molecular mechanism underlying beta-rhizobial symbiosis and identify differences and commonalities between the strategies used by alpha- and beta-rhizobia, we use functional genomic approaches such as transposon sequencing (Tn-Seq), RNA-sequencing, shotgun proteomics and metabolomics (Figure 1c). Our group was the first to determine the whole transcriptome profile of P. phymatum STM815T under nitrogen limitation (a condition that partially mimics the condition encountered by free-living rhizobia in nitrogen-starved soils) and in symbiosis with the economically important papilionoid legume Phaseolus vulgaris (common bean)6. Recently, the combination of metabolomics and dual RNA-sequencing on nodules allowed us to identify novel regulatory pathways in P. phymatum STM815T such as the repression of the production of indole-3-acetic acid (IAA) by the master activator of nitrogen-fixation NifA9.
The goal of this project is to identify and characterize beta-rhizobial genes important for i) bacterial competition in the soil, ii) symbiotic partner recognition, iii) symbiosis establishment, iv) N2-fixation and v) nodule senescence.
The genus Burkholderia accommodates numerous species that colonize a wide range of ecological niches and interact with different hosts [1,2]. These interactions cover a wide spectrum: from mutualism through commensalism to pathogenicity. Some of the species in the genus, such as Burkholderia cenocepacia, are opportunistic pathogens that produce severe infections in cystic fibrosis and immunocompromised patients. Other species such as B. phytofirmans and B. bryophila (now Paraburkholderia phytofirmans and P. bryophila) are mutualists that are beneficial to plants and can be potentially exploited in biotechnological processes. However, their agricultural use as plant growth promoting bacteria (PGPB) is severely restrained due to the potential threat that a few opportunistic pathogenic strains pose to human health.
Good or bad bugs? Friends or foes? The global aim of this project is to unravel the genetic differences leading to mutualism or pathogenicity in Burkholderia and Paraburkholderia species.
In the last years, a lot of effort has been invested in discriminating between the beneficial environmental (the “good”) and the clinical (the “bad”) Burkholderia species [2]. Phylogenetic and taxonomic analyses have given evidence that members of the genus Burkholderia can be separated into two lineages which agree with their different lifestyles [3]. One clade comprises animal and plant pathogens; and the other (recently proposed as the novel genus Paraburkholderia) comprises environmental and plant-beneficial PGPB species.
We hypothesize that the dissimilarities in the genomes of the Burkholderia and Paraburkholderia spp. allow them to interact with different types of hosts, leading to mutualism, opportunism, and/or pathogenicity.
The global aim of this project is to unravel the genetic determinants involved in animal and plant colonization by Burkholderia and Paraburkholderia species using TnSeq-based approaches (Figure 1). TnSeq is a very promising molecular technique that combines both transposon insertional mutagenesis and massive parallel sequencing of the transposon insertion sites to accurately identify genes which are involved in a function of interest in bacteria [4]. This method allows genome-wide screening of the microbial Tn-mutants which are impaired in their relative fitness. It can pinpoint fitness genes and pathways that allow bacteria to competitively survive and proliferate in a given environment. In our study we will use both the classic TnSeq approach [4] and the more recent TnSeq variant called RB-TnSeq5 (randomly barcoded TnSeq) [5], that combines the advantages of TnSeq and rapid quantification of each transposon mutant using unique barcodes that can be amplified by PCR.
The results obtained in this project will lead to a deeper knowledge of the ecology of the genera Burkholderia and Paraburkholderia and will set the steps for the future development of adapted control/antibiotic strategies.
References
[1] Compant et al. 2008. FEMS Microbiol Rev 32(4): 607-626. https://pubmed.ncbi.nlm.nih.gov/18422616/
[2] Eberl et al. 2016. F1000Research, 5: 1007. https://f1000research.com/articles/5-1007
[3] Sawana et al. 2014. Front Genet. 19: 429 https://www.frontiersin.org/articles/10.3389/fgene.2014.00429/full
[4] Opijnen and Camilli. 2013 Nat Rev Microbiol 11: 435-442. https://pubmed.ncbi.nlm.nih.gov/23712350/
[5] Wetmore et al. 2015. mBio 6:e00306-15. https://journals.asm.org/doi/full/10.1128/mBio.00306-15
Funding
This project is funded by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 891542 https://cordis.europa.eu/project/id/891542
Project leader: Dr. Marta Torres
People Involved: Sarah Paszti
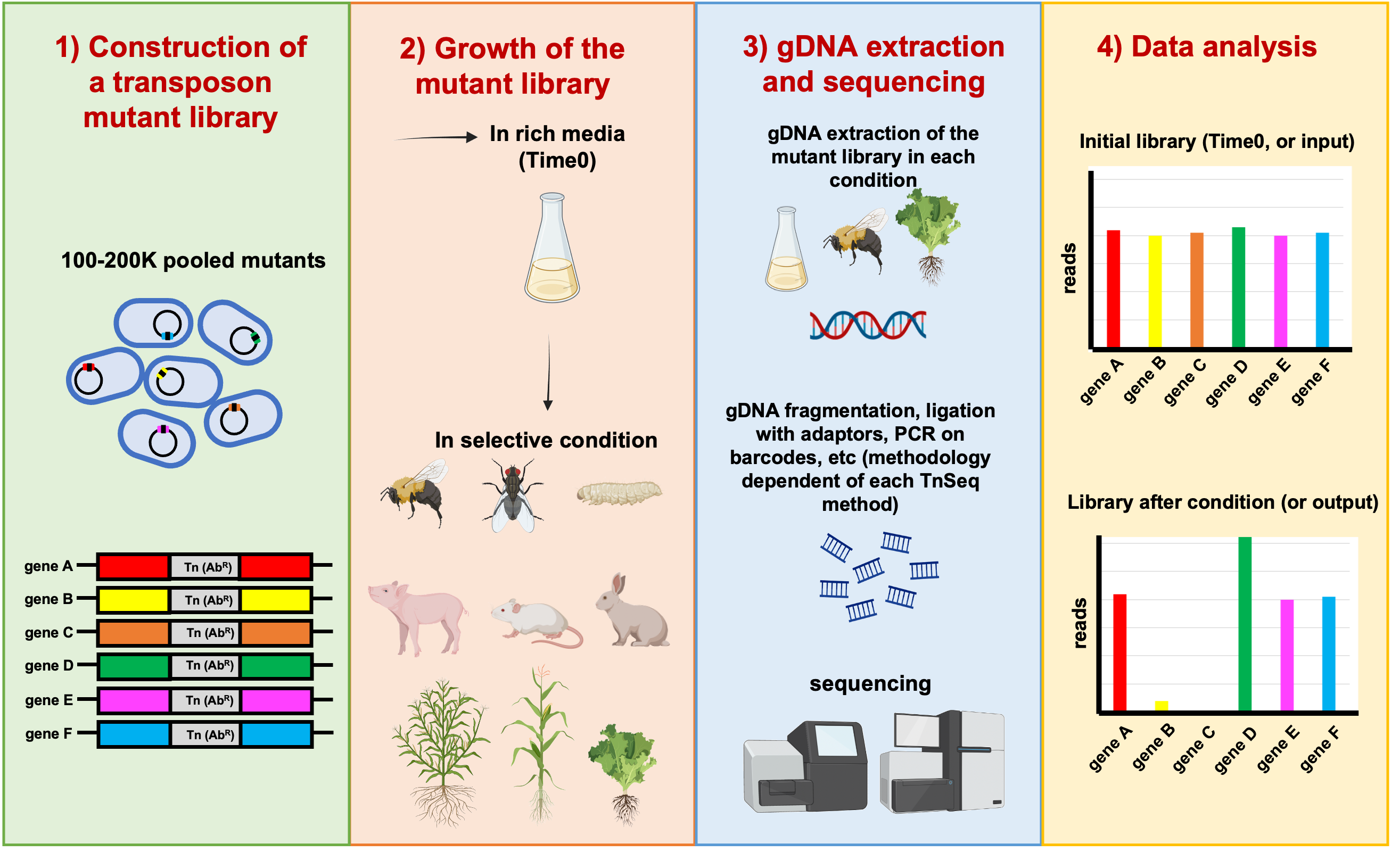
Figure 1: General scheme of the overall TnSeq approach. In this example, genes B and C have reduced fitness; and gene D has enhanced fitness. Copyright: Marta Torres.
In this project we aim to identify colonization factors in different hosts and environments (insects and plants, anoxic conditions), by using the genome-wide mutant profiling method transposon sequencing (Tn-seq). Tn-seq is the combination of transposon mutagenesis with massive parallel sequencing. This powerful approach enables the direct linkage of phenotype and genotype and allows genome-wide analysis. Our strains of interest are Burkholderia cenocepacia H111, a clinical isolate from a cystic fibrosis patient, and Burkholderia thailandensis E264, a soil saprophyte.
Reference
Paszti S, Vitale A, Liu Y, Braunwalder R, Kalawong R, Biner O, Pessi G, Eberl L. Identification of Key Factors for Anoxic Survival of B. cenocepacia H111. Int J Mol Sci. 2022 Apr 20;23(9):4560. https://www.mdpi.com/1422-0067/23/9/4560
Project leader: Prof. Dr. Leo Eberl
People involved: Sarah Paszti, Dr. Marta Torres, Dr. Stefano Gualdi
Collaborations: Dr. Peter Mergaert, University of Paris-Saclay, Dr. Lionel Moulin, University of Montpellier, Annette Vergunst, University of Montpellier
This work is affiliated with the Burkadapt project https://anr.fr/Project-ANR-19-CE20-0007
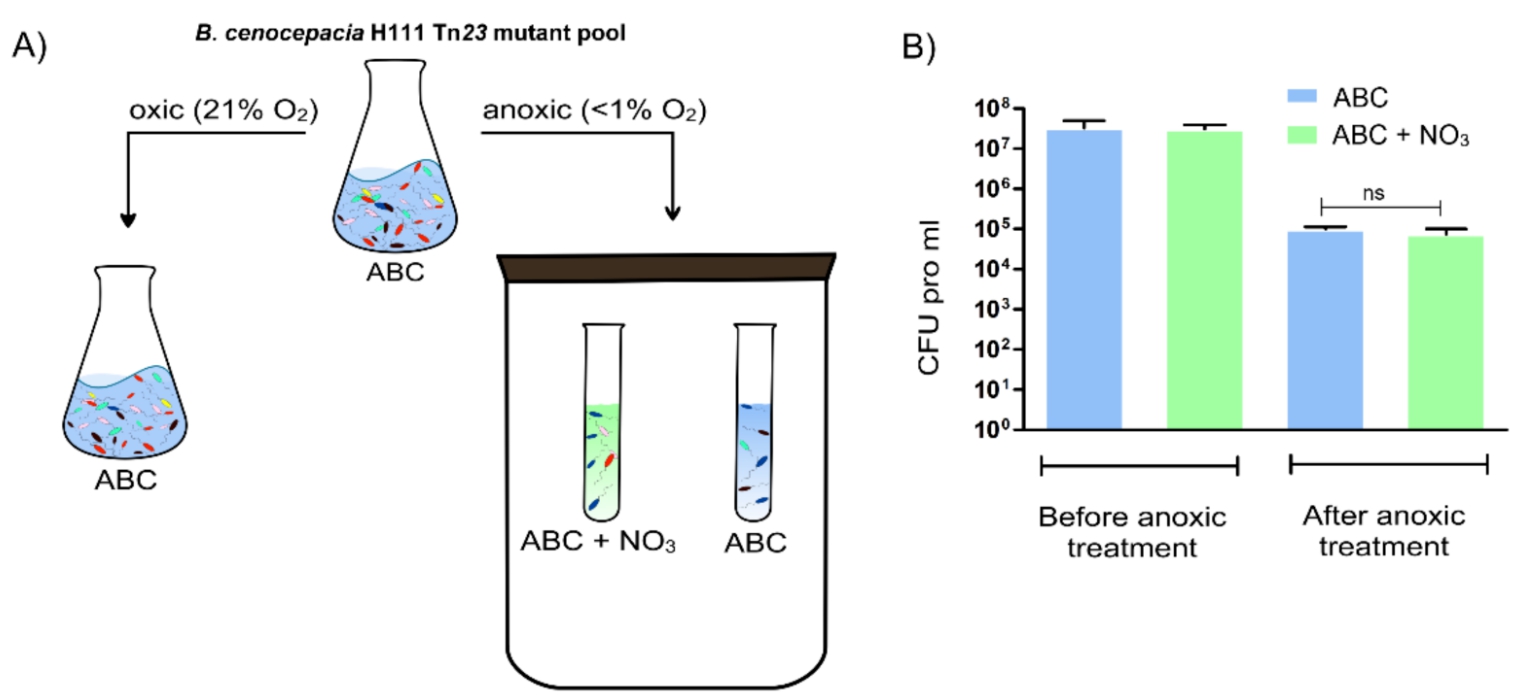
Figure: Overview of the preparation of the three B. cenocepacia H111 samples used for Tn-seq analysis. The control sample (grown aerobically until OD600 of 0.5 at 37 °C in ABC) and the test samples (grown anoxically for 5 days at 37 °C in ABC or ABC with 10 mM NaNO3) were prepared for Tn-seq analysis as described in [1]. B) CFU pro ml at the start and end of the anoxic survival in ABC and ABC with 10 mM NaNO3 for B. cenocepacia H111 wild-type. Error bar = standard deviation (SD), ns = not significant.
The presence of multiple bacterial species coexisting in natural biofilms is the most relevant form of bacterial growth in the environment. Competition for space and resources has triggered the evolution of competitive strategies to persist in such multispecies communities. Our group recently discovered a novel type IVB secretion system (T4BSS) in Pseudomonas sp. strain IsoF, a plant growth promoting rhizobacterium (PGPR), as the first identified T4BSS involved in inter-bacterial killing [1]. IsoF can outcompete numerous proteobacterial species using its T4BSS, as well as provide protection to tomato plants against plant pathogens, such as Ralstonia solanacearum [1].
The aim of this project is to define the killing spectrum of IsoF in mixed biofilms in vitro and as a biocontrol agent in planta, and additionally to characterize its T4BSS. Although we have identified one of this system’s effector-immunity protein pairs, further experiments are required to fully understand the mechanism of action of the T4BSS. Finally, we are also interested in the role of this T4BSS and other genomic islands in root colonization, as well as the potential of this secretion system as a biotechnological tool.
People involved: Miguel Ángel López Carrasco, Dr. Kirsty Agnoli
Reference
[1] Purtschert-Montenegro, G., Cárcamo-Oyarce, G., Pinto-Carbó, M. et al. Pseudomonas putida mediates bacterial killing, biofilm invasion and biocontrol with a type IVB secretion system. Nat Microbiol 7, 1547–1557 (2022). https://doi.org/10.1038/s41564-022-01209-6
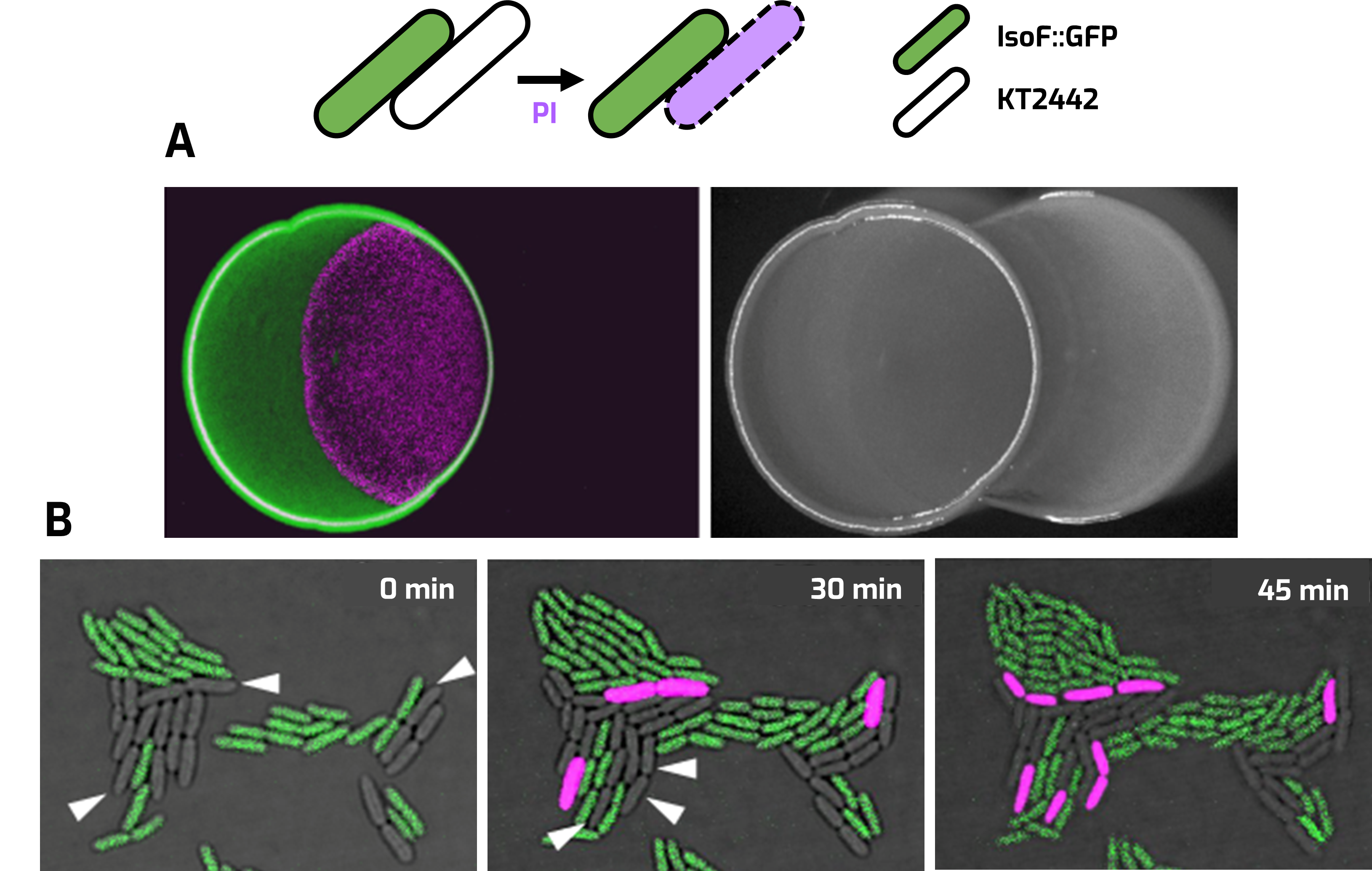
Contact‑dependent killing (CDK) of IsoF::GFP and P. putida strain KT2442. Propidium iodide (PI) staining (magenta) was used for the detection of dead cells. A. Overlapped colonies assay after 24 h. B. Single cell assay under confocal laser-scanning microscopy.
How cell-to-cell signaling in bacteria regulate target genes has been very well studied across species. Some bacteria are multilingual and can master several chemical languages. We previously discovered that Burkholderia sp. strains produce a third signal molecule termed valdiazen which belongs to the unusual diazeniumdiolate class of compounds, that are rarely found in nature [1]. Valdiazen controls the expression of more than 100 genes, indicating that it is a global cellular regulator. Homologs of the valdiazen biosynthesis genes are found in various bacteria, suggesting that valdiazen-like compounds may constitute a new class of signal molecules. We aim to corroborate the role of the valdiazen family of compounds as novel bacterial signal molecules and elucidate the phenotypes controlled by them in a panel of phylogenetically diverse bacteria.
Our recent study identified leudiazen as the second member of the diazenium diolate signal family in the major plant pathogen, Pseudomonas syringae [2]. We demonstrated that leudiazen is a volatile signal that regulates the production of mangotoxin, the major virulence factor in this pathogen. Currently, we aim to characterize the signal and its regulon in several bacteria. This includes structural/functional characterization of mangotoxin and the elucidation of the upstream genetic elements for its production. Valdiazen homologs in other species, such as P. aeruginosa and B. glumae, and the bioactive compounds regulated by the signal are also under investigation.
References
[1]. C. Jenul, S. Sieber, C. Daeppen, A. Mathew, M. Lardi, G. Pessi, D. Hoepfner, M. Neuburger, A. Linden, K. Gademann, L. Eberl, Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 9, (2018).
[2]. Sieber, S., Mathew, A., Jenul, C., et al. (2021) Mitigation of Pseudomonas syringae virulence by signal inactivation. Science Advances. 7 (37)
Project leaders: Prof. Dr. Leo Eberl, Dr. Anugraha Mathew
People involved: Leonardo Paganini, Maria Rodriguez Garcia (PhD students)
Collaborators: Prof. Dr. Karl Gademann and Dr. Simon Sieber (UZH), Prof. Dr. Francisco Cazorla López (University of Malaga)
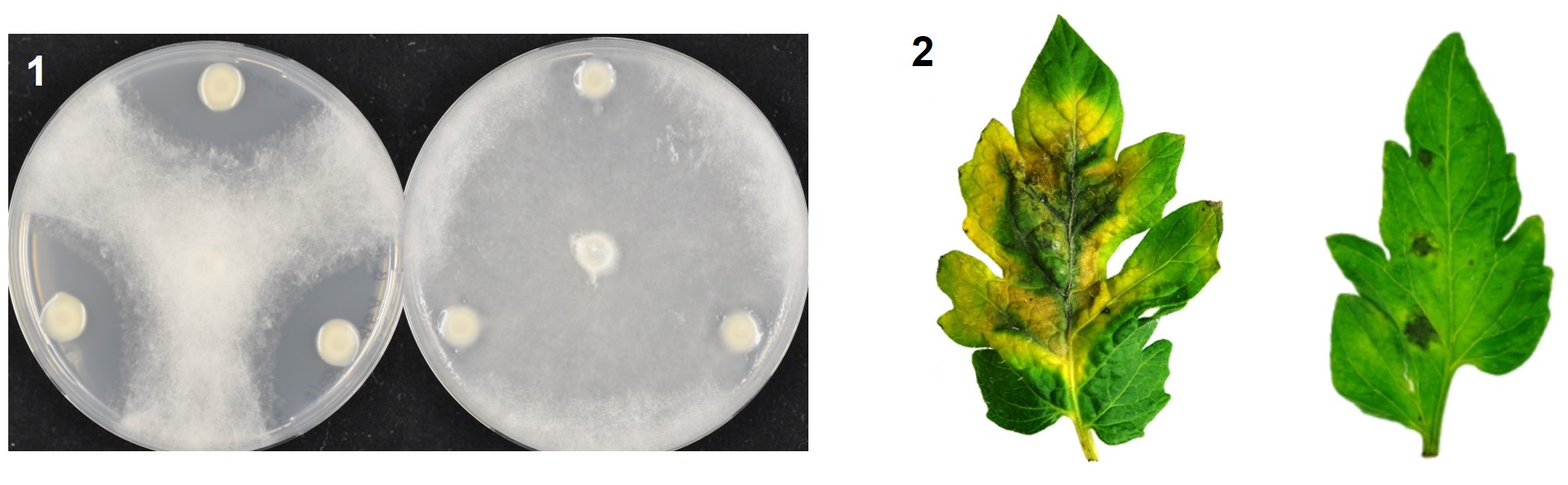
Standard modern farming techniques rely on the heavy use of pesticides to improve crop yields, but many have adverse effects, both to the consumer and to the environment. Bacteria can be used as environmentally friendly alternatives to pesticides, to improve plant growth and to limit plant disease. However, few bacteria have been registered for such plant disease control, and at present none are from the genus Paraburkholderia, which contains talented plant-beneficial bacteria with wide-ranging disease control abilities.
We are currently bioprospecting in the plant microbiome for bacteria that living internally and externally which can protect against important agricultural pathogens, in search of strains that can protect against important agricultural pathogens. We aim to identify new bacterial biocontrol agents, to expand the scope for biocontrol in agriculture.
Project leader: Dr. Kirsty Agnoli
People involved: Dr. Carlotta Fabbri, Dr. Gabriela Purtschert, Anya Schnyder,
Funding: Gebert Rüf Stiftung {https://www.grstiftung.ch/de/media/portfolio~grs-076-19~.html)
Publications:
https://www.mdpi.com/2076-2607/10/12/2480
https://www.nature.com/articles/s41564-022-01209-6

left to right, top to bottom: healthy tomato plants, apple blossom, fluorescent bacterial strain, hypersensitive response assay in apple leaves.
Most bacteria release spherical membrane vesicles (MVs), which are composed of the cellular membrane and are ~20 to 400 nm in diameter. MVs have fundamental roles in various biological processes, including gene transfer, protein exchange, quorum sensing, nutrient acquisition and phage decoying and are involved in bacterial pathogenicity. Because of their various functions, MVs show promise for several applications, such as vaccine development, drug delivery and as scaffolds for enzymatic reactions. Although the significance of the biological functions of MVs is becoming increasingly apparent, how MVs are produced and the factors that influence MV production is not fully understood.
In our group we are studying the functions and formation of MVs. So far, we have discovered that MVs account for the majority of the extracellular matrix proteins in biofilms. This study showed that the protein composition of MVs derived from planktonic and biofilm cells differs, indicating that the content of the MVs reflects the physiology of the cell. Recently, we have identified new pathways and mechanisms for MV formation, triggered by phage related genes in cells under stress, including those in biofilm. We now aim to understand how the different pathways through which MVs are formed influence the content and function of MVs. This knowledge would help us better understand how bacteria respond to the environment and interact with each other.
People involved: Dr. Yi-Chi Chen, Dr. Zaira Heredia, Dr. Ratchara Kalawong
Collaboration: Prof. Dr. Masanori Toyofuku, University of Tsukuba
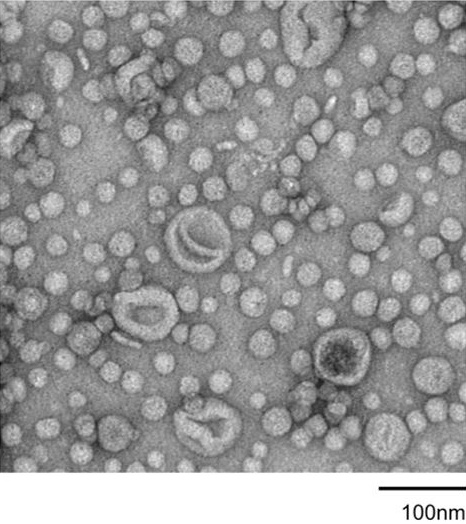
TEM image of MVs (https://doi.org/10.1038/ismej.2017.13)
Lipid bilayers are ubiquitous in all domains of life. Organisms can release small non-replicating particles delimited by a lipid layer into the extracellular medium to fulfill a number of different functions [1]. In the case of bacteria, membrane vesicles (MVs) are spherical bi-layered particles, 20–400nm in size [1, 2]. Depending on their formation pathway, MVs can be classified as different types according to their structure and composition [2].
An increasingly relevant role has been attributed to membrane vesicles (MVs), not only for the host bacterium but also in the interactions between the host and its environment. This project aims to elucidate specific ways in which the contribution of MVs is relevant or provides an advantage to bacterial cultures. The work will be focused on the genus Burkholderia and related species due to their immense potential for secondary metabolite production and also their interactions within the rhizosphere and as human and animal pathogens.
Project Leaders: Prof. Dr. Leo Eberl, Dr. Yi-chi Chen
People involved: Inês Mendes
References
[1] C. Schwechheimer and M. J. Kuehn, “Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions,” Nat Rev Microbiol, vol. 13, no. 10, Art. no. 10, Oct. 2015, doi:10.1038/nrmicro3525.
[2] M. Toyofuku, N. Nomura, and L. Eberl, “Types and origins of bacterial membrane vesicles,” Nat Rev Microbiol, vol. 17, no. 1, pp. 13–24, Jan. 2019, doi: 10.1038/s41579-018-0112-2.
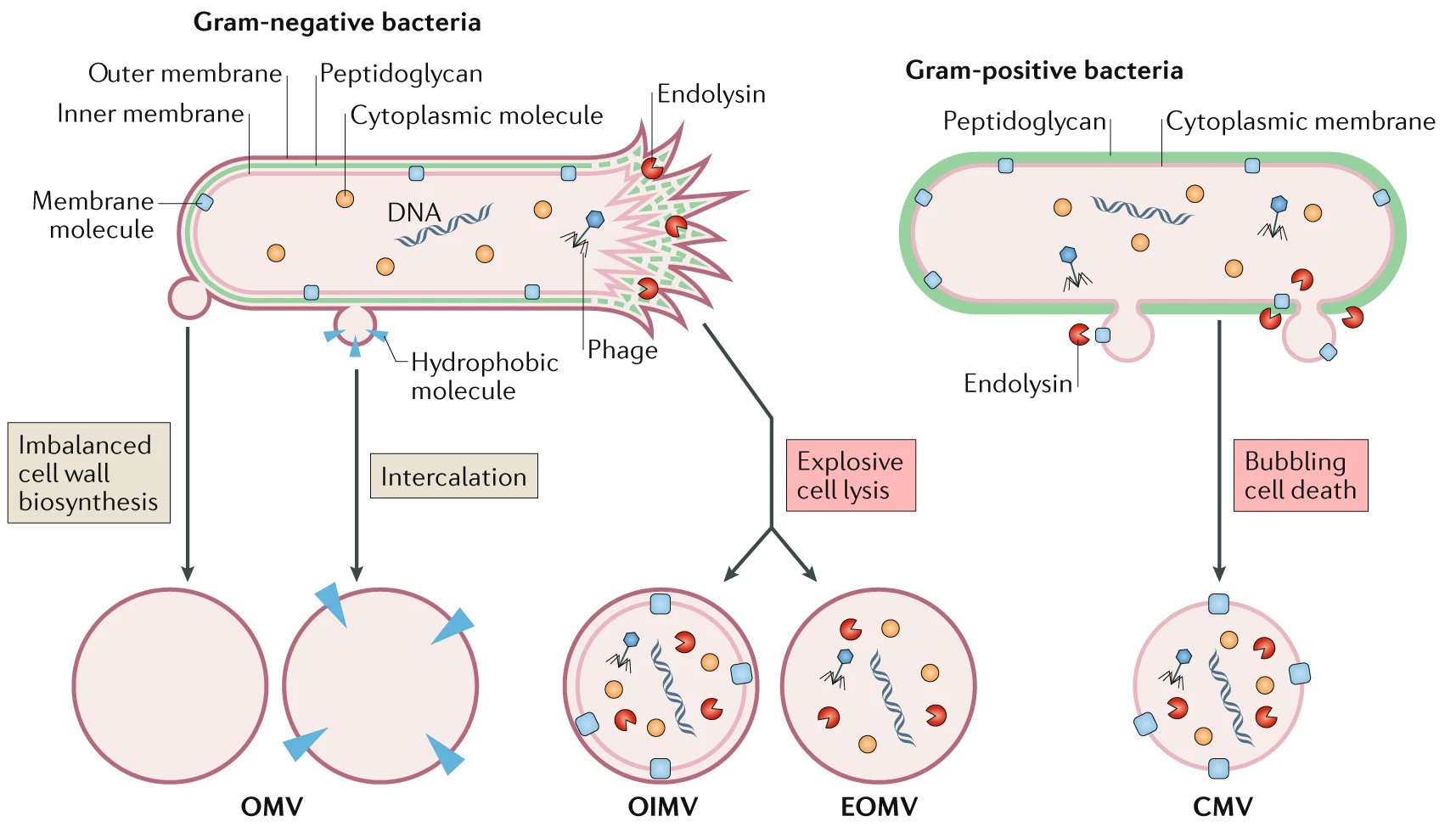
Different routes lead to the formation of distinct membrane vesicle types (https://doi.org/10.1038/s41579-018-0112-2)
This project focuses on unravelling the molecular mechanisms involved in biofilm-associated antimicrobial resistance of the opportunistic pathogen Burkholderia cenocepacia H111.
Biofilms are surface-attached structures composed of bacteria embedded in a self-produced matrix, which is considered an adaptive survival strategy to persist in stressful environments. Biofilms are more than 100-fold more resistant to antibiotics than their planktonic counterparts. We aim to perform a thorough analysis of the mechanisms that biofilm-grown B. cenocepacia cells employ to survive antibiotic treatment. This involves growing Burkholderia in a recently developed biofilm system that allows treatment with different antibiotics. After growth, the biofilms are analysed using a state-of-the-art transposon sequencing technique. This is breaking new ground in understanding the antibiotic resistance mechanisms employed by B. cenocepacia in a biofilm matrix.
People involved: Dr. Stefano Gualdi
Collaboration: Prof. Dr. Tim Tolker-Nielsen, University of Copenhagen
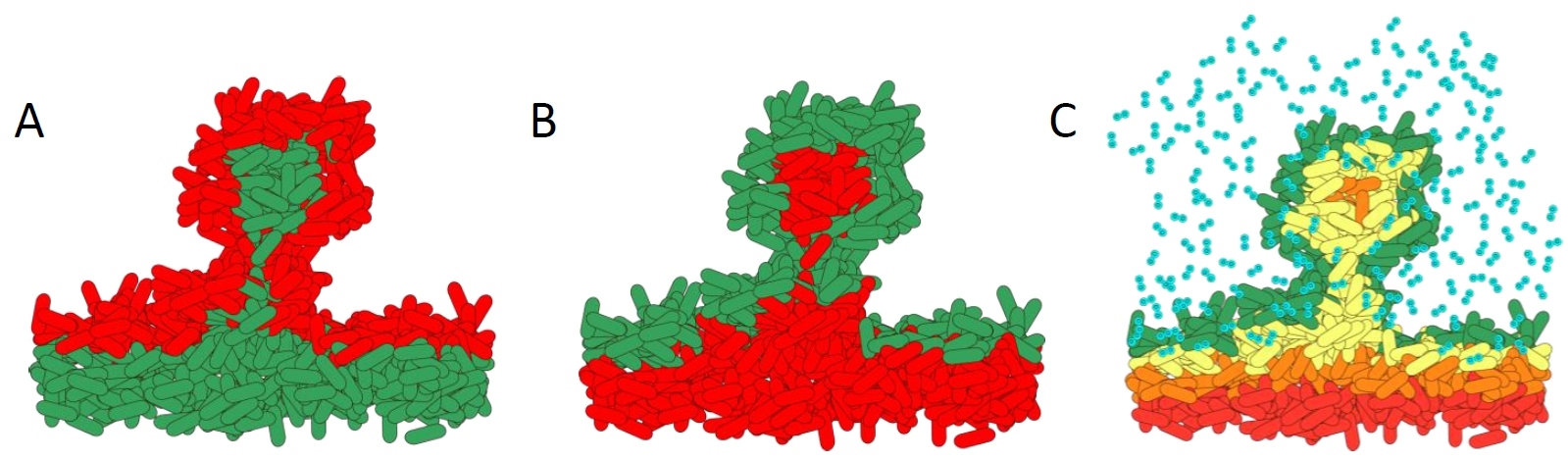
How antibiotics affect bacteria within a biofilm. A). Biofilm treated with standard antibiotics. The metabolically active cells in the top layers of the biofilm are killed. B). Effect of treatment with the novel antibiotic colistin. The metabolically inactive cells are killed as they cannot pump out the colistin. C). Schematic showing the complex microenvironmental conditions within biofilms. The steep oxygen gradients create high growth on the outside layers, whereas the lower layers have slow or no growth.
Diagram courtesy of Prof. Dr. Kasper Nørskov Kragh
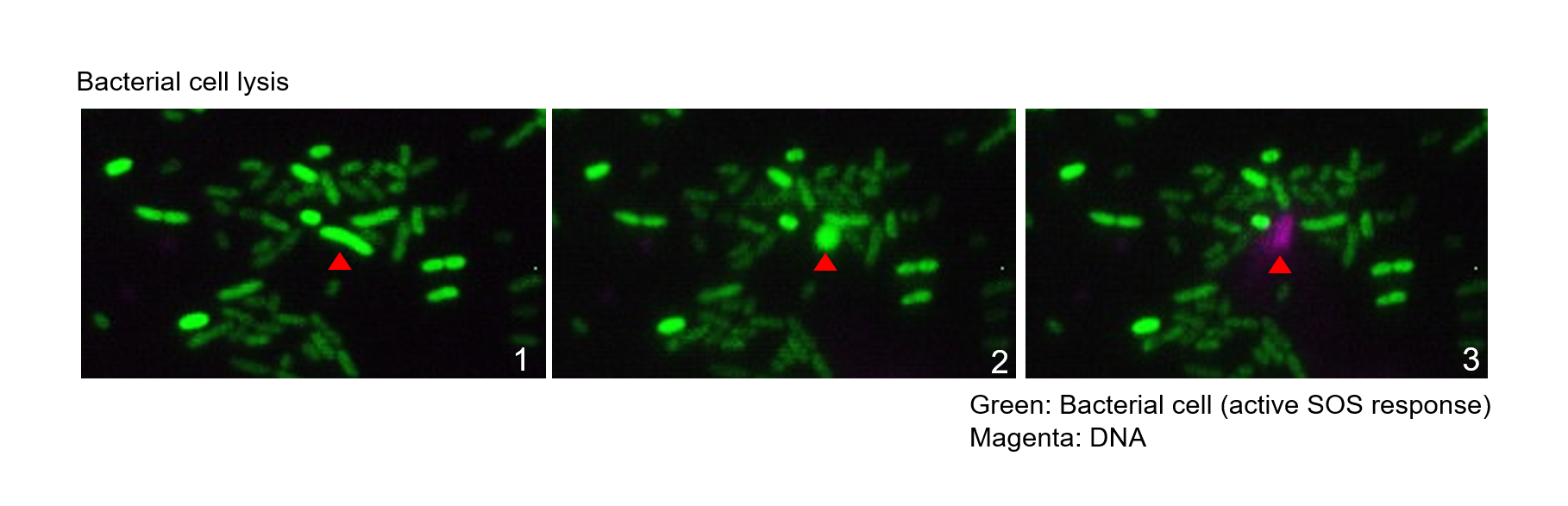
Biofilms are bacterial populations enclosed in an extracellular matrix made of polysaccharides, proteins, DNA and lipids. Biofilm formation is a universal property exhibited by bacteria. Compared to the planktonic lifestyle, biofilms prevail in most habitats on the Earth’s surface and represent the main lifestyle of active bacteria. While biofilms can be beneficial, such as for plant protection and development, they can also be very problematic in the clinic and industry, causing biocorrosion in industrial settings and infections in humans. Biofilms are of particular concern in the clinic, as sessile bacteria show increased resistance to the host immune system and antibiotics. This resistance mostly relies on the extracellular matrix, which acts as a barrier that protects the cells against invasion, predation or immune recognition. The DNA component of the biofilm matrix is hypothesized to result from cell lysis through induction of the bacterial SOS response.This leads to DNA release, which can be enhanced by certain antibiotics. The aim of this project is to define how antibiotics contribute to biofilm formation.
Project leader: Prof. Dr. Leo Eberl
People involved: Dr. Zaira Heredia
Collaboration: Dr. Eleonora Secchi (ETH)