Navigation auf uzh.ch
Navigation auf uzh.ch
Domestication of wheat by early human farmers and the later wheat improvement by scientifically based breeding led to genetic bottlenecks reducing genetic diversity: Thus, modern varieties have lower diversity to cope with biotic and abiotic stresses imposed by pathogen evolution and climate change. Bread wheat landraces, a crop status between wild relatives and elite cultivars, display higher genetic diversity compared to modern breeding material. This broader genetic basis very likely is important for adaptation. To exploit this untapped genetic diversity, we study a diverse collection of 300 Swiss bread wheat landraces (Triticum aestivum L.) within the European Union H2020 project “Activated GEnebank NeTwork” (AGENT, https://www.agent-project.eu) to identify sources of resistance against the wheat powdery mildew disease caused by Blumeria graminis f. sp. tritici which is a yield-reducing fungal pathogen.
To identify novel sources of resistance, a Kmer-based, genome-wide association study (GWAS) was conducted on the landrace panel genotyped with DArTseq technology for seedling resistance using ten powdery mildew isolates with contrasting virulence spectra. We found strong variation of resistance across the Swiss landrace panel. We developed a pan-genome-wide association pipeline based on multiple wheat reference genomes, maximizing the chances of tagging responsible loci for mildew resistance, likely absent in a single reference genome. The association mapping detected multiple known Pm genes like Pm1, Pm2 or Pm4b but also novel regions associated with powdery mildew resistance.
Of note, unlike standard GWAS, our method identifies associations with structural variations and sites not present in a single reference genome, highlighting the relevance of landraces stored at genebanks as a source of novel genetic variation important not only for mildew resistance but any other agronomic trait important for adaptation to a changing environment.
The extent of the AGENT project accross whole Europe will provide material to predict the new resistance loci found in the Swiss collection and ultimately select the best genotypes to use for the next generation of breeding programs.
Funding:
This work is supported by the European Union’s Horizon 2020 research and innovation programme under the grant agreement No 862613 (Activated GEnebank NeTwork [AGENT])
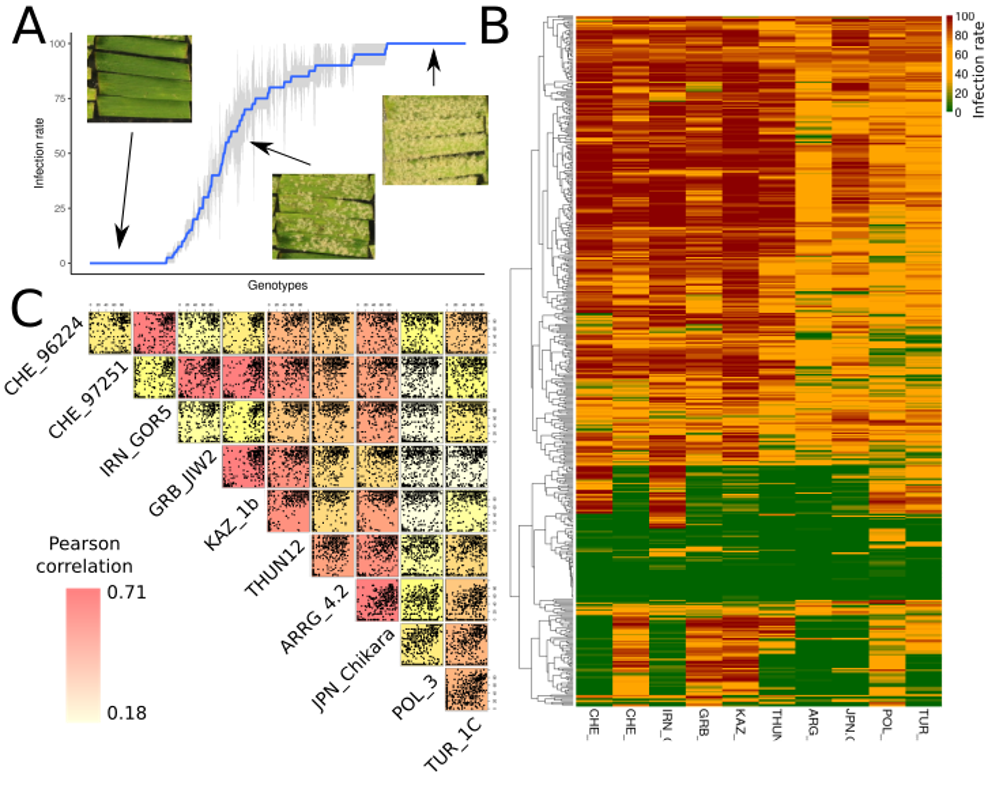
Phenotypic analysis of the Swiss landrace collection: A: resistance pattern of the isolate 96224 across the Swiss collection (pictures show examples for a non-infected, intermediate-infected, and fully infected leaves). B: Heatmap showing all isolates across all accessions of the collection. C: Correlation matrix between all the isolates where every dot represents one accession.
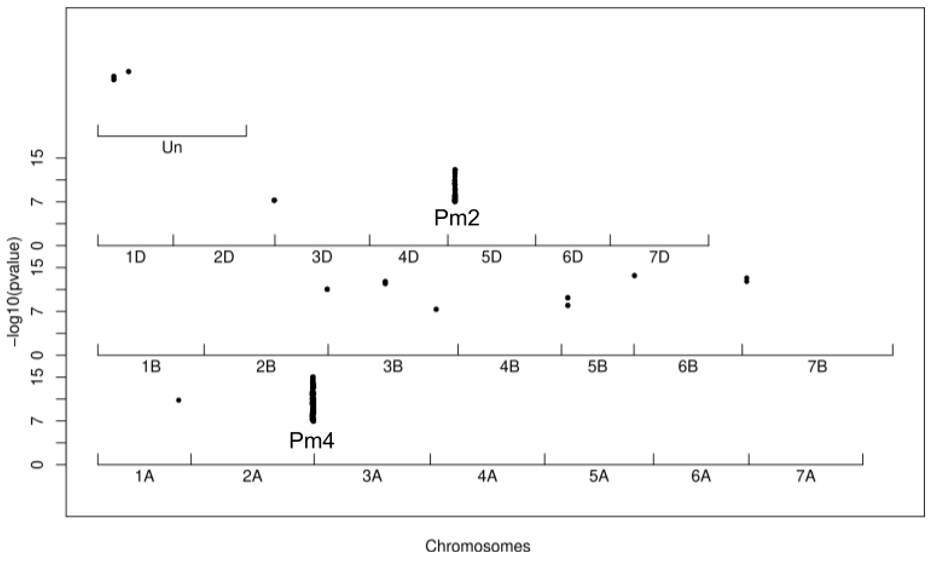
Manhattan plot for the isolate CHE_96224 on the SYMattis genome. Every dot represents a significant kmer mapped onto the SYMattis genome.
The Swiss winter bread wheat cultivar Forno shows near immune levels of leaf rust resistance to all leaf rust isolates tested so far. This durable resistance is controlled by several genes with additive effects, including the race non-specific adult plant resistance genes Lr34 and Lr75, and a race-specific, all-stage resistance gene Lr14a.
Lr34 encodes an ATP-binding cassette (ABC) transporter, and confers durable, albeit partial, resistance to multiple fungal pathogens in the adult plant. Lr75 function as a partial, broad-spectrum adult plant resistance gene but its molecular identity remains unknown. Lr14a encodes a membrane-bound ankyrin repeat protein which has structural similarity with non-selective calcium channels in Arabidopsis and humans. We are interested in studying the underlying molecular mechanisms which result in durable resistance.
Our current research consists of two research lines aiming to understand the molecular mechanism of Lr14a function. One research line focuses on the search for the race-specific AvrLr14a protein which specifically triggers Lr14a-based resistance. Second, we want to characterize a possible calcium channel activity of Lr14a and the downstream resistance signaling pathway using methods and approaches in molecular and cell biology, biochemistry, and proteomics.
Selected papers:
Krattinger et al. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360-1363.
https://www.science.org/doi/10.1126/science.1166453
Singla et al. (2017) Characterization of Lr75: a partial, broad-spectrum leaf rust resistance gene in wheat. Theor Appl Genet 130: 1-12.
https://link.springer.com/article/10.1007/s00122-016-2784-1
Kolodziej et al. (2021) A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nat Commun 12: 956. https://www.nature.com/articles/s41467-020-20777-x
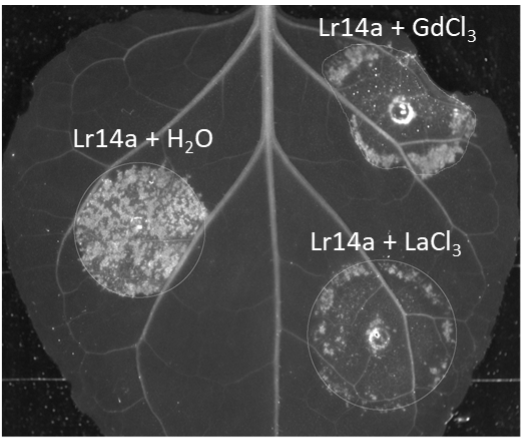 Lr14a induced water-soaking like phenotype in Nicotiana benthamiana afterAgrobacterium infiltration is abolished by calcium channel blockers LaCl3 and GdCl3.
Lr14a induced water-soaking like phenotype in Nicotiana benthamiana afterAgrobacterium infiltration is abolished by calcium channel blockers LaCl3 and GdCl3.In cereals, race-specific resistance (R) genes, including those effective against powdery mildew diseases, are an agronomically important resource for breeding. Race-specific resistance is conditioned by the interaction between an R gene from the host (typically encoding for an NLR receptor), and an avirulence (Avr) gene from the pathogen (typically encoding for an effector). We are interested in understanding the genetic, molecular, and biochemical bases of such resistance in wheat. More specifically, we focus on the interaction between powdery mildew resistance proteins (PM) from wheat and avirulence effector proteins (AVRPM) from the pathogenic fungus Blumeria graminis.
Since many years the focus of research in our lab aims at the molecular identification and functional characterization of Pm genes in wheat. This resulted in the cloning of the NLR encoding race-specific resistance genes Pm2, seventeen alleles of Pm3, Pm8 and Pm17. We have characterized the resistance potential of transgenic wheat lines overexpressing individual Pm genes or combinations of them both under laboratory conditions and in the field. Field trials at the protected experimental field site (www.protectedsite.ch) in Zürich-Reckenholz have shown that overexpression of individual Pm genes or combinations of several genes can increase and broaden their resistance effect.
In recent years we have also become interested in the molecular mechanisms by which NLR receptors recognize the corresponding fungal AVR effectors. Using various techniques such as genetic mapping, genome-wide association studies or effector screens we have since identified multiple avirulence effector genes such as AvrPm1a, AvrPm2, several AvrPm3 effectors (Figure 1), AvrPm8 and AvrPm17 that are recognized by the wheat NLRs Pm1a, Pm2, Pm3, Pm8 or Pm17, respectively and consequently trigger a hypersensitive cell-death response (HR) upon co-expression in the heterologous Nicotiana benthamiana system (Figure 2).We also developed a UV mutagenesis approach to create gain-of-virulence powdery mildew mutants, as a novel tool to identify Avr effectors. Using natural variation, artificial mutagenesis, and domain-swap approaches in avirulence effectors and corresponding NLRs we succeeded in identifying single residues or entire protein subdomains required for HR development or quantitatively contributing to recognition strength.
Genetic analyses in segregating populations allowed us to discover that some AVR-R interactions are controlled by additional pathogen components. We first identified a suppressor of avirulence (SvrPm3), which hinders the recognition of the AvrPm3s by Pm3 through yet unknown mechanisms. We then used UV mutagenesis to unravel other avirulence components in the Pm3/AvrPm3 system. This allowed us to identify a conserved regulator of avirulence and expand our understanding of complex host-pathogen interactions.
Our current research aims at understanding the molecular and biochemical basis of AVR-R recognition and downstream resistance signaling. To investigate the interactions between the different components and their sub-cellular localization, we apply a variety of methods including transient expression through Agrobacterium infiltrations in Nicotiana benthamiana or biolistic bombardment of wheat leaves followed by protein-protein interaction screens (co-immunoprecipitation, split-luciferase, yeast-2-hybrid) and confocal microscopy. Another line of research has been to investigate the host-targets of our avirulence effectors. To do this, we perform large screens using both co-immunoprecipitation coupled to mass-spectrometry (IP-MS) and yeast-2-hybrid libraries.
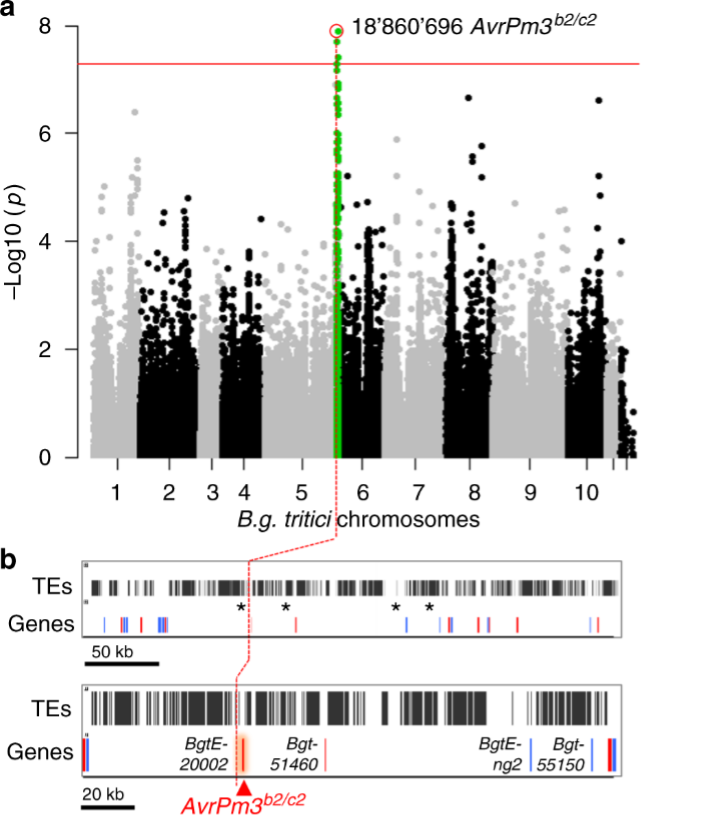
Figure 1. Identification of AvrPm3b2/c2 with a genome-wide association study (GWAS) (Bourras et al., 2019). A group of powdery mildew isolates from a worldwide collection has been used to map a locus associated with avirulence on Pm3b and Pm3c.
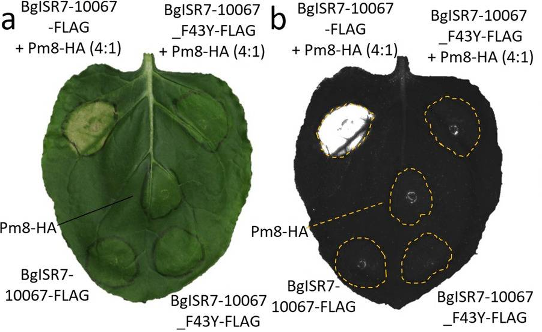
Figure 2. Transient co-expression of Pm8 and AvrPm8 (BgISR7-10067_FLAG) results in hypersensitive response (HR) in Nicotiana benthamiana (Kunz et al., 2023). The virulent form of AvrPm8 (BgISR7-10067_F43Y_FLAG) does not trigger HR when co-expressed with Pm8.
Selected papers
Bourras, S., Kunz, L., Xue, M., Praz, C. R., Müller, M. C., Kälin, C., Schläfli, M., Ackermann, P., Flückiger, S., Parlange, F., Menardo, F., Schaefer, L. K., Ben-David, R., Roffler, S., Oberhaensli, S., Widrig, V., Lindner, S., Isaksson, J., Wicker, T., Yu, D., & Keller, B. (2019). The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nature Communications, 10(1), 1–16. https://doi.org/10.1038/s41467-019-10274-1
Kunz, L., Sotiropoulos, A. G., Graf, J., Razavi, M., Keller, B., & Müller, M. C. (2023). The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen. BMC Biology, 21(29). https://doi.org/10.1186/s12915-023-01513-5
We want to understand the molecular basis underlying the mode of action of race-specific resistance (R) genes in wheat. Most of the R genes cloned to date encode intracellular NLR (nucleotide-binding domain leucine-rich repeat) receptors. In recent years new types of R genes were identified. Among them are genes encoding tandem kinases, which occur in various domain combinations. In addition to the tandem kinase encoding WTK4 (Gaurav et al. 2022) we have cloned Pm4, a non-canonical gene conferring race-specific resistance against wheat powdery mildew (Blumeria graminis f. sp. tritici, Bgt). Pm4 encodes two alternatively spliced protein variants: Pm4b_V1 and Pm4b_V2. The Pm4 proteins have a unique domain architecture, including a serine-threonine kinase (S_TKc) domain, multiple C2-domains and transmembrane region(s) (MCTP). We showed that both protein isoforms are required in wheat to confer resistance (Figure A and B) (Sánchez-Martín et al. 2021).
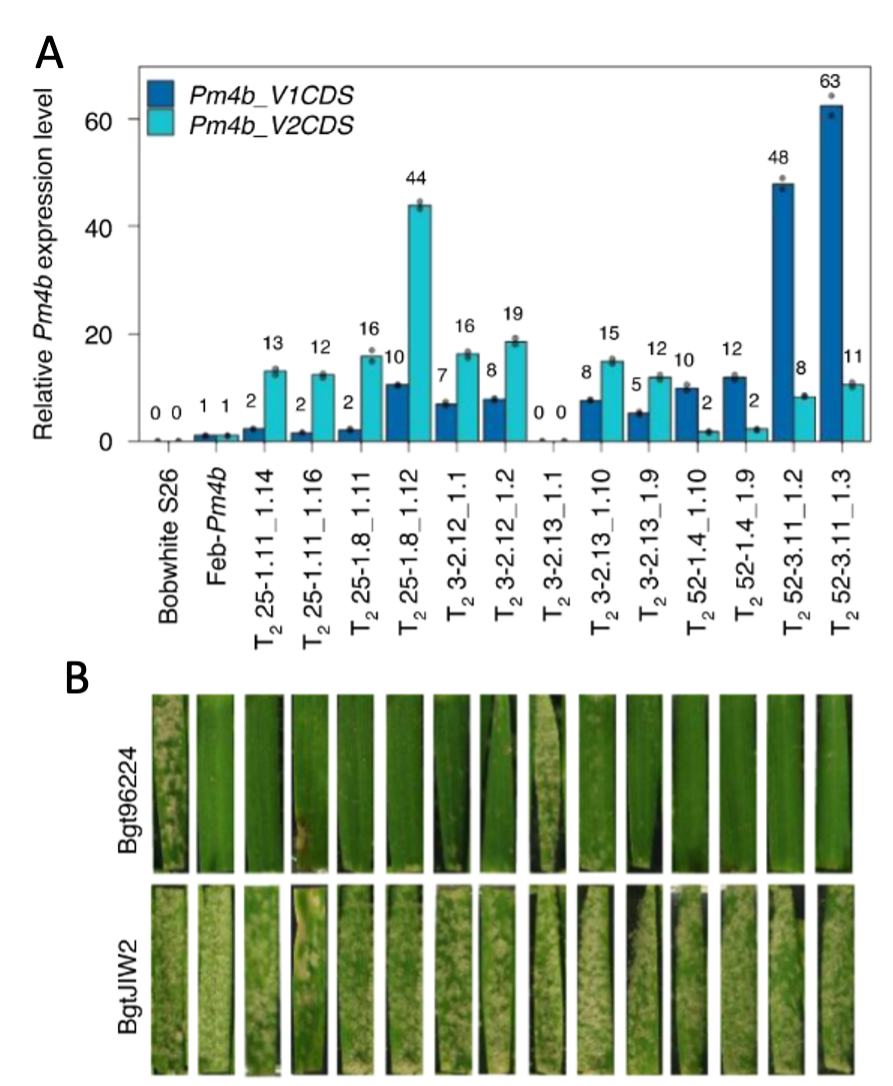
(A) shows Pm4b_V1 and Pm4b_V2 RNA expression levels in transgenic Bobwhite S26. Except for line T23-3 .2.13_1.1 all transgenic lines show expression of both Pm4b splice variants. As expected, the control Bobwhite S26 does not show Pm4b expression.
(B) shows leaf segments from the respective plants illustrated in panel A, which were then infected either with Pm4b avirulent Bgt96224 or Pm4b virulent BgtJIW2. The transgenic lines expressing both Pm4b splice variants show race-specific resistance against Bgt96224.
(Sánchez-Martín et al. 2021)
We aim at understanding the molecular and biochemical basis of non-canonical R-AVR recognition and the down-stream signalling leading to resistance for the wheat WTK4 tandem kinase as well as for Pm4. To address this question, we rely on a multidisciplinary approach based on techniques in molecular and cell biology, biochemistry and proteomics. These include for example transgenic wheat plants, wheat protoplasts, transient protein expression in Nicotiana benthamiana, protein-protein interaction assays (IP-MS, yeast-two-hybrid screens) and kinase assays. We also aim at identifying the AVRs recognized by WTK4 and Pm4.